SOFTWARE
 GenePattern
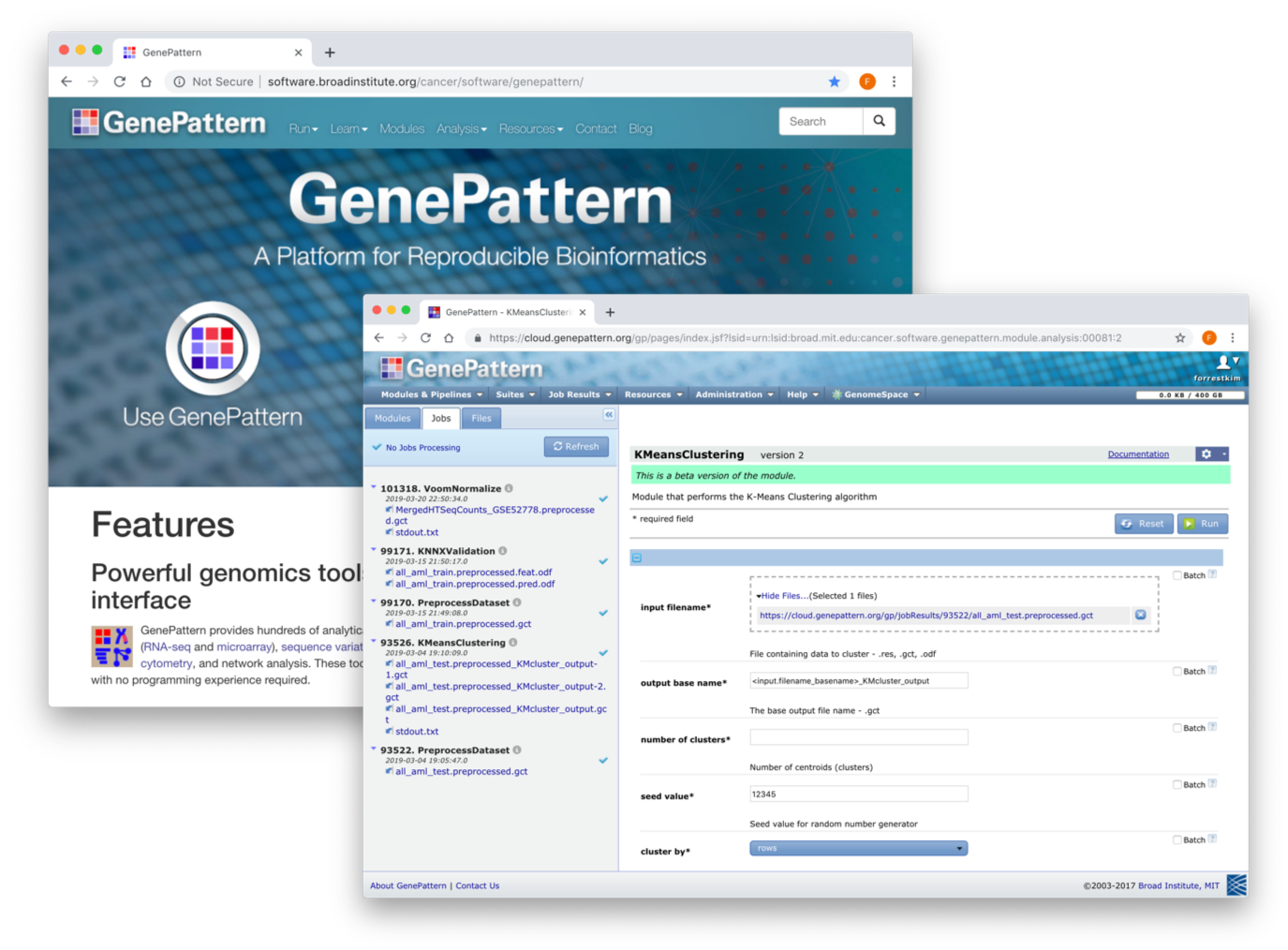
GenePattern
We have developed a software package called GenePattern, which provides a comprehensive environment that can support a broad community of users at all levels of computational experience and sophistication, access to a repository of analytic and visualization tools and easy creation of complex analytic methods from them and the rapid development and dissemination of new methods. Perhaps the most important feature of GenePattern is that it supports a mechanism to guarantee the capture and independent replication of published computational methods and in silico results.
 GenePattern Notebook
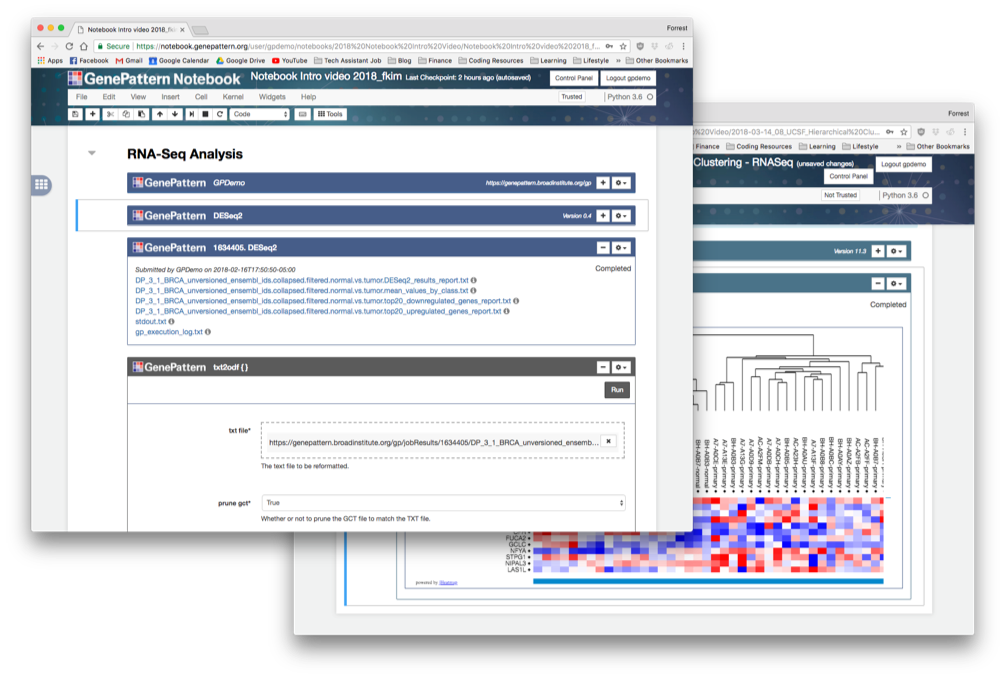
GenePattern Notebook
Interactive analysis notebook environments promise to streamline genomics research through interleaving text, multimedia, and executable code into unified, sharable, reproducible “research narratives.” However, current notebook systems require programming knowledge, limiting their wider adoption by the research community. We have developed the GenePattern Notebook environment, to our knowledge the first system to integrate the dynamic capabilities of notebook systems with an investigator-focused, easy-to-use interface that provides access to hundreds of genomic tools without the need to write code.
 IGV
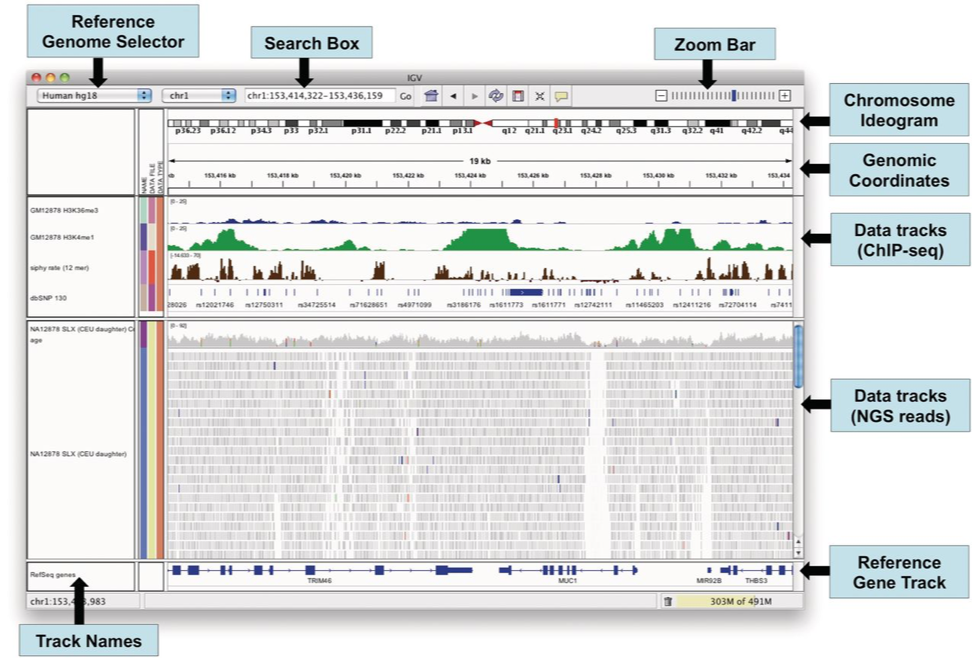
IGV
The Integrative Genomics Viewer (IGV) is a high-performance visualization tool for interactive exploration of large, integrated genomic datasets. It supports a wide variety of data types, including array-based and next-generation sequence data, and genomic annotations.
 (1)
(1)
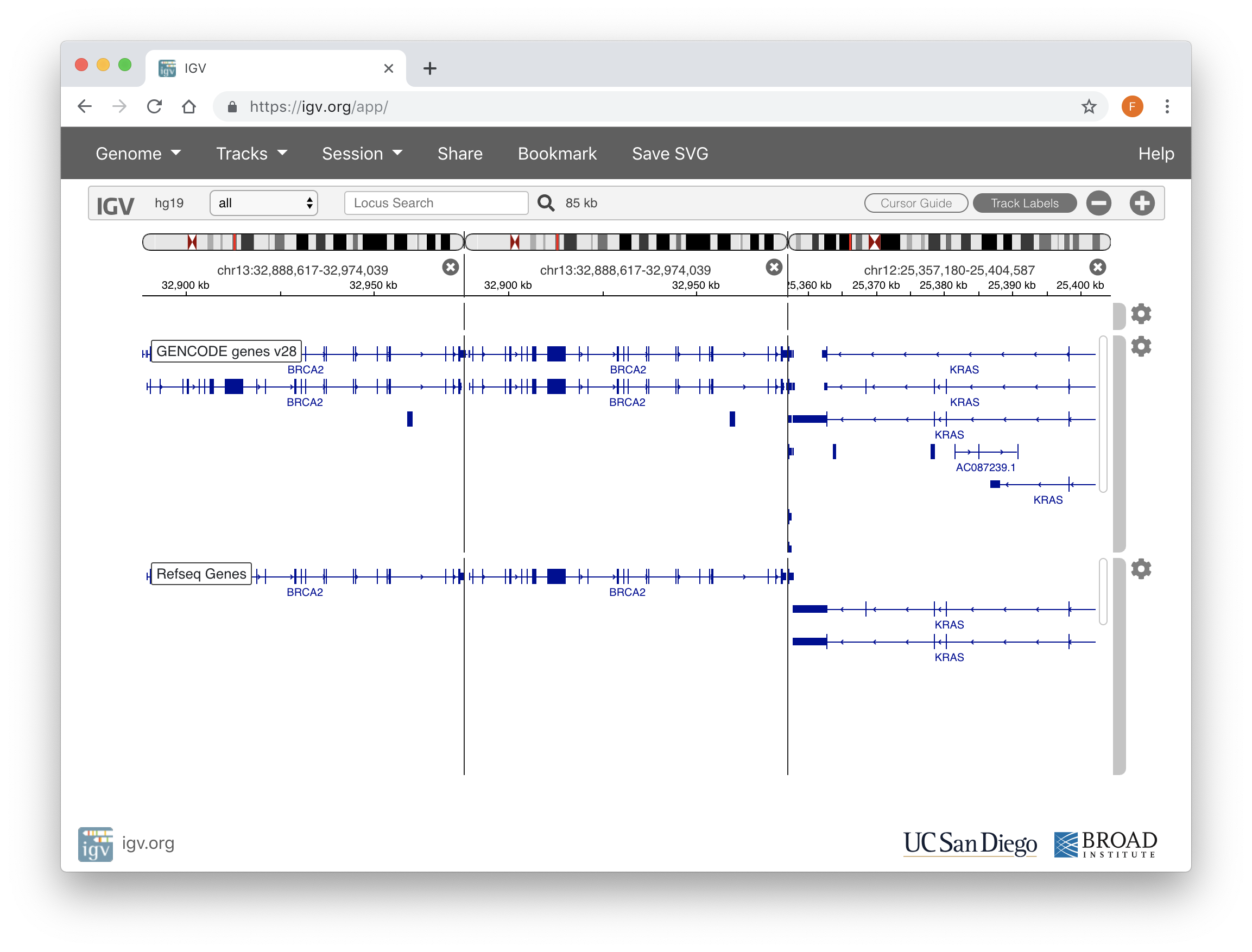
 IGV.js
IGV.js
IGV.js is an embeddable interactive genome visualization component written in JavaScript and CSS. It is based on the Integrative Genomics Viewer(IGV), and developed by the same team.
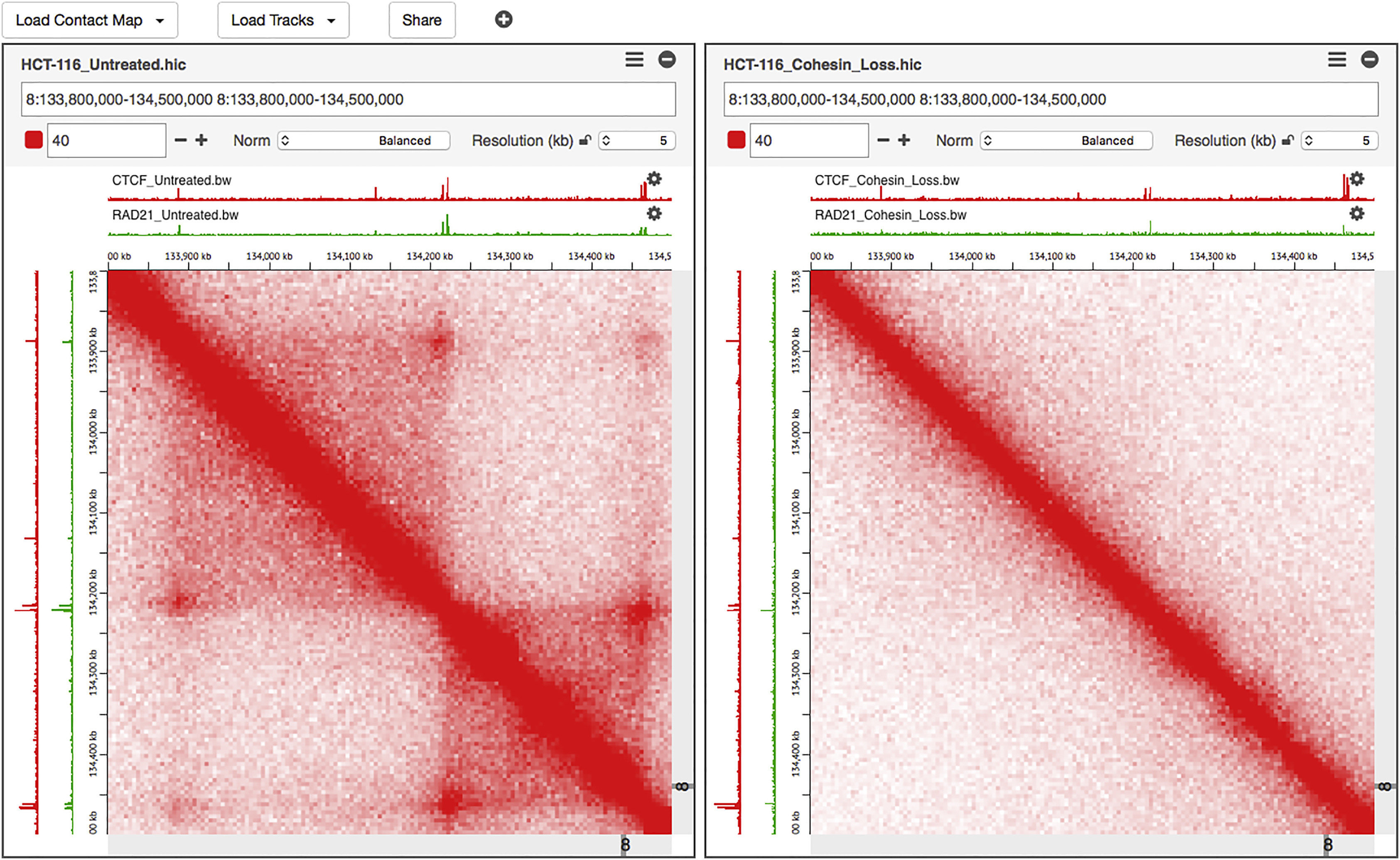
 Juicebox.js
Juicebox.js
Juicebox.js is an interactive contact map viewer for .hic files, written in JavaScript and CSS. It is based on the desktop Juicebox visualization application, and developed by the same team.
 (2)
(2)
 (3)
(3)
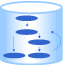
 GSEA
GSEA
Gene Set Enrichment Analysis (GSEA) is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states (e.g. phenotypes).
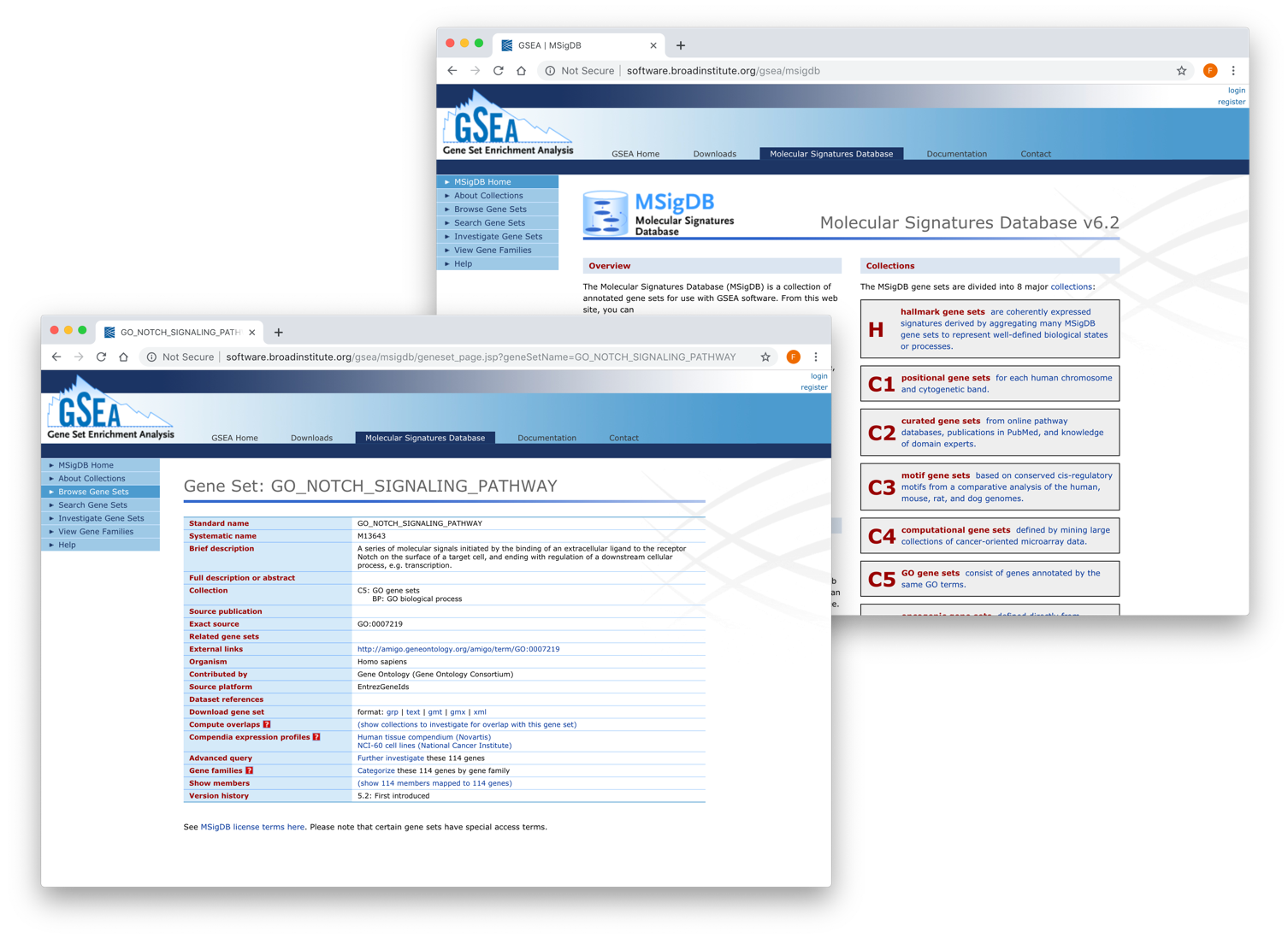
 MSigDB
MSigDB
The Molecular Signatures Database (MSigDB) is a collection of annotated gene sets for use with GSEA software. From this website, you can search for gene sets by keyword, browse gene sets by name or collection, examine a gene set and its annotations, download gene sets, compute overlaps between your gene set and gene sets in MSigDB, Categorize members of a gene set by gene families, and view the expression profile of a gene set in a provided public expression compendia.
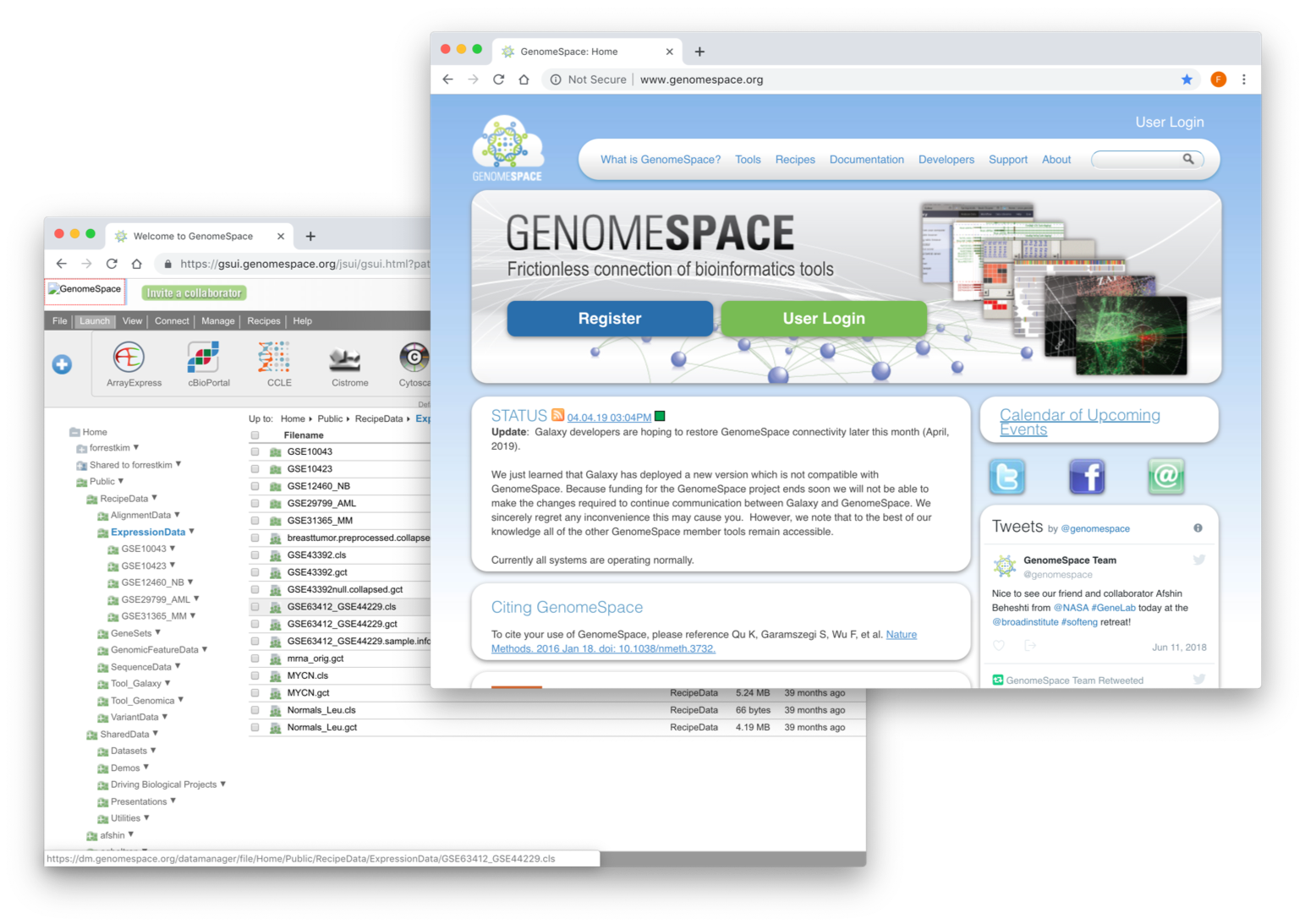
 GenomeSpace
GenomeSpace
GenomeSpace is a cloud-based interoperability framework to support integrative genomics analysis through an easy-to-use Web interface. GenomeSpace provides access to a diverse range of bioinformatics tools, and bridges the gaps between the tools, making it easy to leverage the available analyses and visualizations in each of them. The tools retain their native look and feel, with GenomeSpace providing frictionless conduits between them through a lightweight interoperability layer. GenomeSpace does not perform any analyses itself; these are done within the member tools wherever they live – desktop, Web service, cloud, in-house server, etc. Rather, GenomeSpace provides tool selection and launch capabilities, and acts as a data highway automatically reformatting data as required when results move from the output of one tool to input for the next.
- Helga Thorvaldsdóttir, James T. Robinson, Jill P. Mesirov. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics 14, 178-192 (2013).
- James T. Robinson, Douglass Turner, Neva C. Durand, Helga Thorvaldsdóttir, Jill P. Mesirov, Erez Lieberman Aiden. Juicebox.js Provides a Cloud-Based Visualization System for Hi-C Data. Cell Systems (2018).
- http:/../software.broadinstitute.org/gsea/index.jsp